Advancements and challenges in One Health
Barbara Roda1, Alessandra Bònoli2, Vittorio Sambri3, Maria Careri4
1Dipartimento di Chimica “Giacomo Ciamician”, Alma Mater Studiorum – Università di Bologna; 2Dipartimento di Ingegneria Civile, Chimica, Ambientale e dei Materiali, Alma Mater Studiorum – Università di Bologna; 3DIMEC – University of Bologna and Unit of Microbiology, The Greater Romagna Hub Laboratory; 4Dipartimento di Scienze Chimiche, della Vita e della Sostenibilità Ambientale, Università di Parma
Contribution presented by Aldo Roda
Abstract
The One Health concept emphasizes the interconnectedness of human, animal, and environmental health, advocating for a comprehensive and collaborative approach. Since its inception, various global interventions have addressed complex health challenges such as epidemics and pandemics. In developed countries, numerous collaborative platforms have been established, adopting international strategies to tackle both local and global health issues. Current challenges in One Health include endemic and emerging zoonotic diseases, food safety and security, antimicrobial resistance, and wildlife diseases. Developing new bioanalytical techniques and methods based on biosensors, ‘omics’ approaches, and high-throughput analytical techniques that are rapid, low-cost, and capable of providing robust quantitative results is crucial.
Keywords
One Health, Environmental impacts assessment, Microbiology, Exposome, Stressor, Bioanalytical strategies, Diagnostics, Biosensors, Omics.
© Roda, Bònoli, Sambri, Careri, 2024 / Doi: 10.30682/annalesps2402g
This is an open access article distributed under the terms of the CC BY 4.0 license
1. Introduction
One Health is a holistic approach that recognizes the interconnection between humans, animals, and ecosystems to address health challenges and issues. Transdisciplinary and cross-sectoral collaboration is an essential component of the One Health approach to effectively address complex health challenges [1, 2] (Fig. 1)1.
Already in the nineteenth century, physician Rudolf Virchow argued that “between animal and human medicine, there are no dividing lines, nor should there be” [3]. This established the foundation of the concept “one medicine, one pathology, one health” and the basis of the model organism approach to human disease research [4]. One of the most significant effects of this approach is the development of collaborative platforms by the World Health Organization (WHO), Food and Agriculture Organization of the United Nations (FAO), and Organizzazione mondiale della sanità animale (OIE), such as the Global Early Warning System for Major Animal Diseases Including Zoonoses, the OIE/FAO Network of Expertise on Avian Influenza, and the FAO/OIE Network of Expertise on Avian Influenza Crisis Management Center for Animal Health. A significant policy shift occurred in late 2008 when the WHO, FAO, and OIE, together with the United Nations Children’s Fund (UNICEF), the United Nations Influenza System Coordination (UNSIC) and the World Bank, jointly approved the “One World, One Health” policy framework (OWOH) [5].
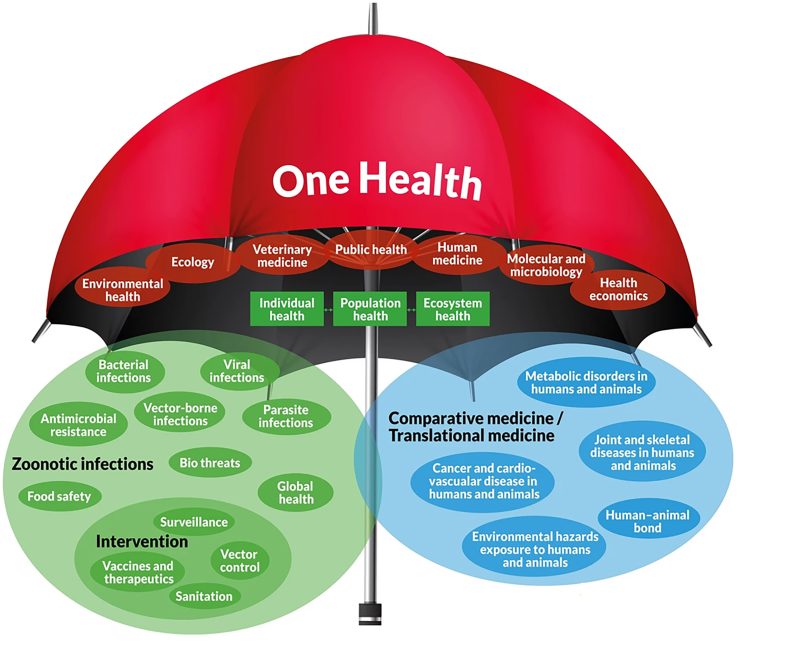
Fig. 1. The One Health umbrella concept. Reproduced via license CC BY 4.0 from [2].
In recent years, the concept of “One Health” has been implemented to fully consider the complexities and interconnectedness of different influences that have impacts at a larger system level, including in addition to human and wildlife health, the equally inseparable ecosystem health. Through wider application, the concept developed into what is now called “One Health”, which is currently widely recognized as an integrated and unifying approach focusing on interactions among people, animals, plants and the environment to holistically address health threats [6].
Overall, the connections between the health of humans, animals, plants, and their shared environment need thorough investigation. This interconnection becomes closer as expanding human populations come into closer contact with wild animals, climate change promotes the spread of zoonotic diseases and new epidemics, travel and world trade lead to an increase of the rapid diffusion of diseases at the global level.
The “One Health” concept considers all potential connections between environmental exposure and health outcomes, and how this subsequently influences public health and other environmental processes. It encompasses chemical fate and transport in the environment, environmental degradation of materials, and how exposure may impact ecosystem processes and human health. This perspective aligns with green-chemistry principles [7] and full–life-cycle assessment [8]. However, current epidemiological studies often focus primarily on a single human or environmental stressor or a single group of stressors, thus failing to effectively manage the combinations and interactions between multiple environmental exposures [9].
The complex context of real-world exposures highlights the limitations of targeted methods for investigating the exposure-disease relationship. Instead, assessing the cumulative and interactive effects of multiple stressors should be a top-priority issue in exposure studies. In this context, cutting-edge analytical and bioanalytical-based techniques through the development of new methods and workflows to assay the exposome – the totality of exposures encountered throughout the life course – hold great promise for the future of exposomics by exploiting new tools such as high-throughput metabolomics [10]. Including metabolomics or other omics profiling within the exposome enables a systems-biology framework to study the effects of exposures, leading to a more comprehensive understanding of events at the molecular scale needed to gain insights into potential toxicity mechanisms at the population level [11].
Currently, high-resolution mass spectrometry (HR-MS) coupled with liquid chromatography (LC) has increasingly been accepted as the predominant platform for the global analysis of the organic chemical exposome. MS-based metabolomics methods, both targeted and untargeted, should be applied together to understand the complexity of organic chemical exposures throughout the life cycle of organisms. The different ways in which HR metabolomics can be applied to study the human exposome can be considered in linear cause-effect relationships, such as (1) associating environmental exposure data to respective plasma levels of a chemical or its metabolite; (2) associating plasma levels of an environmental chemical to related metabolite levels and pathway effects; (3) associating dose-response effects in model systems with human dose-response relationships; (4) integrating exposure-associated metabolic effects with disease phenotypic markers; and (5) using an integrated omics strategy to identify pathophysiologic responses [12]. Advances in bioinformatic tools and big-data analytics strategies for biological and environmental monitoring of exposure for organic chemicals can be fully utilized in future research. Sampling, sample treatment, measurement, and data analysis using computational tools and artificial intelligence all need to be tuned to each other to obtain robust information, taking into account that to assess the risk of chemical exposure it is important to also investigate the transformation products of environmental pollutants. In this regard, examining in depth the transport and transformation processes affecting chemicals in the environment and the human body at increasingly lower concentrations poses new challenges for bioanalytical methods [13]. Integration of high-resolution mass spectrometry with bioanalytical methods, particularly in vitro bioassays to assay the exposome, can enable improved prediction of in vivo response from in vitro bioassay results, thus providing information to assess the risks arising from the effects of all chemicals present in a sample. “Big data” methods could help clarify the relationship between signals and toxicity according to the general goal of exposure studies to capture stressors as much as possible to reflect the holistic impact: a new way of identifying mixtures in disease in the epidemiological setting has to be promised to arrive at an integrated exposure assessment.
In recent decades, inorganic toxicity has also raised concerns for the environment, human and animal health, with reference to emerging exposure to elemental toxicants [9], even though in the field of animal health, the use of biomarkers is less developed than in human health, in a One Health perspective, animal biomonitoring can provide important information on the interfaces between humans, animals and the ecosystems, supporting the management of health risks. Therefore, a transfer of knowledge from human biomonitoring to farm or wild animals is essential in a farm-to-human risk assessment framework. Identification of the presence of non-essential elements in living organisms, in addition to essential elements with multi-element analysis and in-depth knowledge of their functions, is urgently required [14]. The emerging field of metallomics, aimed at unraveling the molecular mechanisms of metal-dependent life processes, is essential for studying biomolecule-metal ion interactions within organisms and ecosystems, as well as the distributions and functions of toxic elements in environmental samples and biota. Inductively coupled plasma mass spectrometry (ICP-MS) has emerged as a powerful tool for determining the inorganic chemical exposome [8]. Additionally, its high sensitivity, elemental selectivity, and multiplex potential have made ICP-MS–based immunoassay a fundamental technique for bioanalyte quantification after element-tagged immunoassay [15].
Lastly, successful implementation examples of the One Health approach are also emerging in the area of nanotoxicology. Man-made nanoparticles present unprecedented persistent exposure through air pollution due to global development in nanoscience and nanotechnology [16]. Likewise, the use of nanomaterials such as nanopowders, nanofibres, nanocapsules, metal nanoparticles, and metal oxides in the food industry raises the issue of oral exposure to nanomaterials introduced into food supply systems. Recent studies have revealed knowledge gaps in the toxicological properties of nanomaterials and insufficient understanding of their interactions with biological systems [17]. Once again, the One Health approach could offer a comprehensive approach to improving food safety and public health outcomes. Furthermore, a One Health approach could create the conditions for comparative nanotoxicology to progress rapidly with reference to engineered nanomaterials before a safe-by-design approach can prove successful.
In this article, we describe the advancements and challenges in One Health in identifying emerging risks in the environment, farming, and animal breeding, also associated with climate change. The assessment of environmental impacts will concern water, air, and soil. Attention will be paid to how climate change, global trading, and massive movements of large masses of people have changed the epidemiology of various infectious diseases, as well as to the relevance of the One Health approach in promoting the prevention and control of diseases transmitted between animals and humans, in combatting antimicrobial resistance, in ensuring food safety, in preventing environmental threats to human and animal health and in addressing many other challenges. This article also aims to highlight innovative approaches based on biosensors and related bioanalytical tools, as well as One Health findings.
2. Environmental-impacts assessment in water, air, soil. A One Health approach in farming
The One Health approach is a cross-disciplinary strategy designed to improve human health at the human-animal-environment interface. It aims to design and implement programs, policies, legislation, and research where multiple sectors communicate and collaborate to achieve better public health outcomes (WHO Regional Office for Europe, 2021). In defining the environment, it encompasses the complex interactions of physical, chemical, and biotic factors, including land, air, water, soil, and all living things that interact within it. This includes natural ecosystems and areas transformed by humans such as urban, agricultural, and productive regions. The health of the environment determines its condition and its ability to function at its best.
Environmental-impact assessment
Several methods can be employed to evaluate the state of the environment and its quality, while also taking into account the pressures imposed by anthropic activities.
The carbon footprint, usually measured in equivalent tons of CO2, corresponds to the whole amount of greenhouse gases (GHG) produced to support a person’s lifestyle and activities, both directly and indirectly [18]. According to the WHO, a carbon footprint is a measure of the impact of activities on the amount of carbon dioxide produced, including direct emissions, as well as emissions required to produce the electricity associated with goods and services consumed.
Ecological Footprint accounting measures the demand on and supply of nature (Global Footprint Network, https://www.footprintnetwork.org). On the demand side, the Ecological Footprint sums up all productive areas for which a population, person, or product competes, measuring the ecological assets required to produce the natural resources it consumes (including plant-based food, fiber products, livestock, and fish products) and to absorb its waste, particularly carbon emissions. On the supply side, a city, state, or nation’s biocapacity represents the productivity of its ecological assets, including cropland, grazing land, forest land, fishing grounds, and built-up land. Life Cycle Assessment (LCA) is a process for evaluating the environmental burdens associated with a product or process by identifying and quantifying energy and materials used, and wastes released into the environment [19].
Environmental impacts in farming: air emissions, water use, wastewater and manure, energy
Intensive farming has significant negative effects on the environment, including air pollution, energy and water consumption, wastewater and solid waste management, and contributions to global warming. These issues have severe consequences for all living things: animals, humans, and plants. According to the LCA approach, some specific impact categories, such as GHG emissions, water quality, biodiversity, fertility and soil quality, and Human and Environmental Toxicity can be identified in order to quantify and assess the environmental impact.
Airborne emissions
The agro-livestock industry contributes substantially to airborne pollution through both direct and indirect emissions and, in general, plays a role in degrading air quality by generating nitrogen compounds primarily from manure, volatile organic compounds other than methane (NMVOC), carbon black (BC), heavy metals, dioxins, and particulates, including PM10 and PM2.5, originating from various farm-level activities
Cattle are the main source of greenhouse gas (GHG) emissions from the livestock industry globally, accounting for 65% (4.6 Gt CO2 equivalent) of sector emissions annually, of which 20% come from the production of milk. Livestock systems are accountable for indirect emissions resulting from land-use change, energy consumption, fertilizer use, and transportation emissions associated with livestock operations and supply chains, in addition to direct emissions. The three primary sources of greenhouse gas emissions from the milk industry are enteric fermentation (CH4), manure management (CH4 and N2O), and feed production, processing, and transportation (CO2 and N2O) emissions, which together account for more than half of all emissions [20].
Water supply and consumption
According to the FAO, around 69% of all renewable freshwater (rivers, lakes, and groundwater) is used for irrigation, livestock, and aquaculture [21]. Risks associated with climate change and water management represent an absolute priority. Effective choices to reduce the water impact of animal production include using low-water-consumption cultivation, implementing more efficient crop irrigation, and reducing human consumption of animal products. The water footprint is emerging as a crucial indicator of sustainability in the agricultural and food sectors. It measures the total volume of freshwater used to produce the goods and services consumed by an individual or community, or produced by a business. Dairy production and agriculture are highly water-intensive, with high variability depending on the region, type of crops, and irrigation methods, which account for up to 90% of water withdrawn from reservoirs. Key water-consuming activities in breeding include animal watering, stable hygiene, air conditioning, and spraying water on animals to increase thermal comfort.
Wastewater and manure management
Animal manure contains nitrogen in the form of complex compounds that enter the nitrogen cycle through bacterial degradation when spread on land. Under suitable conditions, nitrous oxide, a GHG with a high global-warming potential, is produced. Manure also contains other organic compounds such as carbohydrates and proteins, which are naturally converted by aerobic or anaerobic bacteria into carbon dioxide and methane, respectively. To mitigate the environmental and health impacts associated with wastewater [22] and manure, several structural intervention technologies can be implemented throughout the process (Fig. 2). These include reducing the surface area affected by effluents, frequent manure removal, renewal of litter boxes, decreasing air velocity and temperature above effluents, and treating exhausted air through ammonia scrubbing. Solid-liquid separation can reduce the solid content of sewage percolating into the soil when distributed, positively affecting the reduction of ammonia and odor emissions. Storing sewage in specific basins can reduce airborne and odor emissions, and the presence of insects and pathogens. Organically fixed nitrogen becomes a useful fertilizer, significantly enhancing the fertilizing properties and agronomic value of slurry per surface unit. Livestock manure, combined with other organic waste, can be transformed into biogas through anaerobic digestion.

Fig. 2. Wastewater components and their effects. Loosely based on a figure in [22].
The phase of agronomic distribution of effluents and the management of agricultural land are critical for nitrogen emissions. Ammonia emissions occur during and immediately after the distribution phase. Effective emission mitigation strategies, guided by the principle of minimizing soil surface/air exchange and reducing the effluent’s exposure time to the air, can include optimizing the use of effluents by selecting the appropriate spreading time when crop uptake is highest. This can be further enhanced by adopting precision farming techniques, ensuring homogeneous distribution in the field, reducing overlaps, increasing working width, and promoting a more uniform crop response.
A LCA model can be employed to evaluate the environmental impacts of farming. This model facilitates the identification of the most significant impacts related to processes, energy and fuel consumption, water use, and supplied products, such as animal feed and plastic packaging, solid waste, wastewater, and greenhouse gas emissions. Notably, the most significant impact factors are often associated with the farm’s internal use of fossil fuels for operations, the sourcing of animal feed, and circular packaging [23] (Fig. 3).
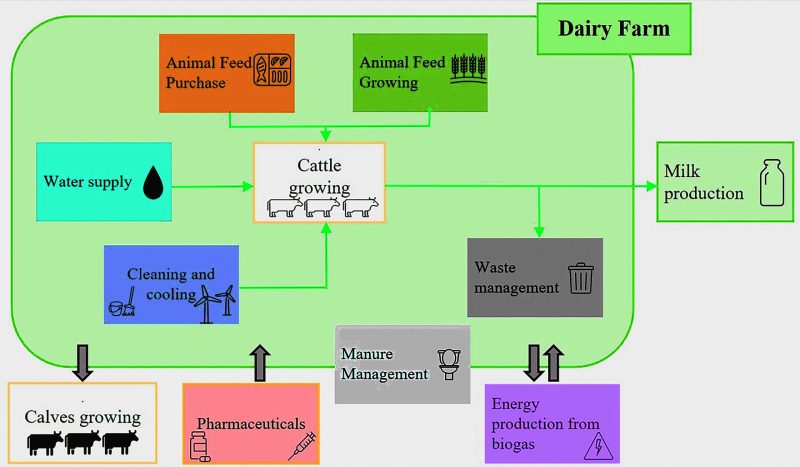
Fig. 3. A conceptual model flow chart for a dairy farm. Reproduced via license CC BY 4.0 from [23].
In this regard, identifying the key factors that affect dairy farm management is the essential first step towards increasing awareness and evaluating the outcomes of initiatives focused on One Health and sustainability.
3. One Health perspective: Microbiology and beyond
It is now well known that climate changes, global trading, and the large-scale movement of people have altered the epidemiology of many infectious diseases, including those caused by the so-called arboviruses (ArBo viruses). This group of viruses shares the characteristic of being transmitted between animals and humans through the bites of hematophagous arthropods, primarily mosquitoes and ticks. This brief paper highlights some of the most recent outbreaks of ArBo infections that have occurred outside tropical areas, particularly in Europe and Italy over the past two decades.
Chikungunya and Dengue
Chikungunya (CHIKV) is a mosquito-transmitted virus belonging to the Togaviridae family. In tropical areas, the main vector for CHIKV is Aedes aegypti [24]. Since the last decade of the 20th century, CHIKV has spread eastward from its original location in Africa, causing significant epidemic outbreaks in the Indian Ocean islands, such as La Réunion (a French territory), and in the Indian subcontinent. In 2007, an unexpected outbreak of “summer fever” began in a rural area near Ravenna and Cesena in Emilia-Romagna, close to the Adriatic coast. This outbreak was quickly identified as CHIKV infections. However, a key question arose: which vector was responsible for this outbreak, given that A. aegypti was not reported in the affected area? The most prevalent mosquito species in this region was A. albopictus (the Asian Tiger Mosquito), previously considered to have negligible vector capability for CHIK. A. albopictus was first identified in Northern Italy in the 1990s and, within approximately 20 years, became the most prevalent mosquito in Italy. The investigation into the 2007 CHIKV epidemic adopted a comprehensive One Health (OH) approach, including the collection of mosquitoes using traps in the affected areas. This rapidly led to the identification of CHIKV RNA in mosquito pools. Sequencing investigations revealed a genetic modification of CHIKV in Italy, specifically a single amino acid substitution in the E1 protein (A226V), which increased the vector capability of A. albopictus for CHIKV [25]. This explained the virus’s spread in Italy. This was the first outbreak of CHIKV outside tropical regions. In subsequent years, CHIKV has been shown to spread locally in various parts of Europe, causing outbreaks with varying numbers of patients. The most recent significant outbreak occurred in 2017 in Lazio [26], with over 800 suspected and confirmed cases in Rome. In this instance, the prompt identification of the circulating virus (genetically similar to those of the Indian clade identified 10 years earlier in Romagna, suggesting a similar geographical origin) was achieved through OH approaches, substantially contributing to the rapid and effective control of the epidemic. Dengue virus (DENV) is an ArBo flavivirus, primarily transmitted by A. aegypti in both sylvatic and urban cycles. In the past year, over 80 cases of local autochthonous transmission of DENV have been reported in Lombardy and Lazio. In this outbreak, A. albopictus was identified as the main vector, suggesting an adaptation process of DENV to this locally prevalent Italian mosquito [27].
West Nile Virus
West Nile Virus (WNV), a member of the Flaviviridae family, is maintained in nature through a cycle involving mosquito vectors (mainly of the genus Culex) and birds [28]. WNV was originally identified in the 1950s in Uganda’s West Nile region. Since its appearance in New York City in 1999, the infection had been largely overlooked. The first cases of WNV infection in Italy were reported in the 1990s in horses in the Fucecchio area (Tuscany) [29]. No further cases were reported in Italy until 2008, when a patient with WNV meningoencephalitis was found in the Bologna area [30]. The epidemiology of WNV is heavily influenced by climatic factors such as rainfall and temperature, which promote mosquito vector concentration in the environment [31]. One of the major threats posed by WNV, apart from cases of central-nervous-system involvement (which generally have a high mortality rate), is the potential transmission of WNV from asymptomatic infected individuals via blood transfusions and organ donations. To mitigate this risk, an integrated nationwide surveillance plan has been implemented in recent years. This plan, enabled by the OH integration system, considers data from vectors, birds, horses, and humans. When WNV RNA is first detected (usually in mosquitoes trapped in rural environments), routine screening for WNV in blood and organ donations begins [32].
4. The role of bioanalytical chemistry in the One Health approach
Bioanalytical sciences hold great promise in the One Health approach, enabling the simultaneous monitoring of various parameters across human, animal, and ecosystem health. The integration of both simple and advanced techniques, such as separation and hyphenated techniques, mass spectrometry, ligand-binding assays, biosensors, miniaturization devices, and data science, has facilitated the development of highly sensitive and reliable detection and quantification methods. These high-functionality tools play a crucial role in bridging the diverse aspects of the One Health concept.
4.1 Biosensors in the One Health approach
Biosensors represent a significant technological advancement in the One Health management, offering improved quality control in the food industry, environmental monitoring, and healthcare. With higher sensitivity and specificity compared to traditional diagnostic methods, biosensors provide rapid results in a portable format. They utilize biological agents as receptors, including enzymes, whole cells, nucleic acids (deoxyribonucleic acid, DNA, or riboNucleic acid, RNA), and monoclonal antibodies, to interact with target analytes. A transducer converts the biochemical response from these recognition elements into a detectable output proportional to the analyte concentration (Fig. 4).
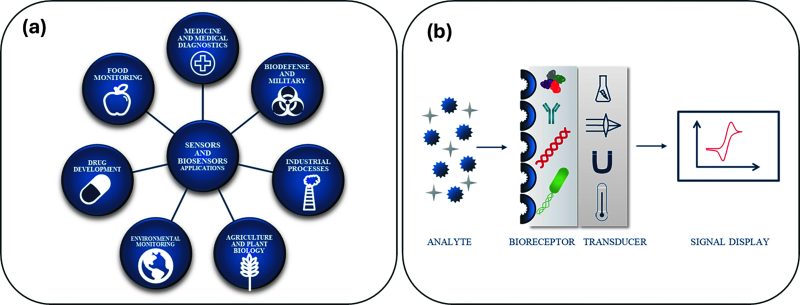
Fig. 4. (a) Biosensor applications; (b) Scheme of a biosensor. Reproduced via license CC BY 4.0 from [33].
Innovative biosensors aim to develop a new generation of environmental and health monitoring systems. The combination of transducer functionality, advanced design, and the miniaturization of readout electronics plays a crucial role in the development of wearable devices for real-time health monitoring. The increasing research and industrial interest in new medical biosensors integrated into point-of-care (POC) systems, alongside the portability of Internet of Things (IoT)-based devices have achieved considerable progress thanks to the pandemic-induced digital health transformation [34].
This article will focus on two broad areas of application of these devices: molecular and clinical diagnostics; monitoring environmental pollutants that bio-magnify within food chains, and a critical evaluation on how they can improve human health.
Biosensors for molecular and clinical diagnostics
Portable biosensors are ideal platforms for realizing minimally invasive diagnostic tools able to provide molecular-level information, to be used for implementation of personalized medicine. In diagnostics, portable glucose sensors have revolutionized diabetes care by enabling patients to measure their blood glucose levels independently, without the need for centralized lab testing. The current state of global health is critically concerning, particularly due to the recent COVID-19 pandemic. With the emergence of new health threats, it is essential to have tools that can accurately detect, diagnose, and monitor diseases [35]. Diagnostic tests often rely on bioassays that detect clinically relevant biomarkers, such as proteins, antibodies, and nucleic acids. Depending on their type, biomarkers can offer information related to prevention, diagnosis, prognosis, or therapeutic response. A major challenge in biomarker detection is their typically low concentration, especially in the early stages of a disease. Significant efforts are still being invested in enhancing the signal generated when these biomarkers bind to a sensor’s capture surface. In this context, the One Health approach benefits from the use of highly sensitive biosensing tools and devices. through the implementation of magnetic beads in electrochemical binding assays [36] also in magneto-electrochemical platforms for tumor biomarker detection, as devised by Careri et al. [37]. Biosensors capable of detecting multiple biomarkers offer simple, reliable, and effective systems to address various aspects of diagnosis and disease treatment comprehensively. Roda et al. developed an ultrasensitive dual chemiluminescence immunosensor to rapidly determine SARS-CoV-2 Immunoglobulin A (IgA)- and Immunoglobulin G (IgG)-specific immunoglobulins in saliva and serum, facilitating the non-invasive monitoring of early immune responses to a disease. This approach also investigates the diagnostic and prognostic utility of salivary IgA for large-scale screening to assess vaccine efficacy [38].
Biosensors can also identify, monitor, and track infectious diseases, including zoonoses. Scientists have used aptamer-based biosensors to detect Salmonella in slaughterhouses. Additionally, biosensors can help address antimicrobial resistance, a significant public health threat caused by the misuse of antibiotics in human and animal healthcare, by detecting antibiotic-resistant bacteria in various specimens from animals, humans, and the environment [39]. Furthermore, biosensors can detect volatile organic compounds in animal breath, such as ketones and ethanol, which indicate high blood glucose levels [40]. This capability is useful for diagnosing diseases like foot and mouth disease, bovine tuberculosis, and brucellosis. Wearable sensors have drastically transformed health monitoring and engagement [41].These devices, which started as simple tools for tracking basic physical activity, have evolved into advanced systems capable of molecular sensing through less invasive biological fluids such as skin interstitial fluid (ISF), sweat, tears, and saliva. Applications include early-stage diagnosis through the detection of disease-specific biomarkers within the skin compartment, such as in melanoma, or the monitoring of biological reactions like urinary infections or microbial biofilms in wounds, without needing to remove dressings, thus reducing the risk of contamination. The use of wearable sensors in veterinary medicine and agriculture holds significant potential for enhancing diagnostics and management practices. However, adapting these sensor technologies from human to animal and plant systems requires consideration of species-specific physiological factors and environmental variables. Thus, developing robust sensor platforms for continuous monitoring in dynamic agricultural environments remains a significant challenge.
Biosensors for environmental monitoring
Biosensors are essential tools for monitoring soil, water, and air samples to detect pollutants resulting from the accumulation of harmful substances, such as pesticides, potentially toxic elements, pathogens, toxins, and endocrine-disrupting chemical compounds. These pollutants are often released due to increased industrial activities, rapid urbanization, and population growth. One notable feature of biosensors is their capability for continuous in-field monitoring of various pollutants. Pollutants are widely spread in the air, soil, and waters, affecting all living systems, especially human health. Within the One Health approach, environmental pollution monitoring needs for rapid, robust, reliable, cost-effective, and portable biosensing devices capable of detecting pollutants from complex matrices without requiring prior sample preparation. Compared to biomedical biosensors, those designed for environmental monitoring face additional challenges due to the complexity of ecological matrices, which can interfere with pollutant recognition. Additionally, finding sustainable solutions for environmental monitoring is critical, as controlling toxic substances is fundamental to pollution remediation. To this end, effective analytical strategies have been proposed to develop highly sensitive and selective advanced biosensors to detect trace organic and inorganic pollutants Examples include immunosensors based on antibodies fixed on interdigitated array microelectrodes for detecting pesticide residues, and enzyme-based biosensors for determining heavy metals in river water and phosphate in soil [42].
4.2 “Omics” technologies in the One Health approach
“Omics” encompasses a broad spectrum of analytical approaches, including genomics, proteomics, transcriptomics, metabolomics, and metallomics. These technologies utilize advanced high-throughput methods, data analysis, and bioinformatics to study the roles, interactions, and functions of various analytes within biological systems. Combining multi-omics studies allows for holistic insights into the comprehensive impacts of several factors and different target analytes (Fig. 5).
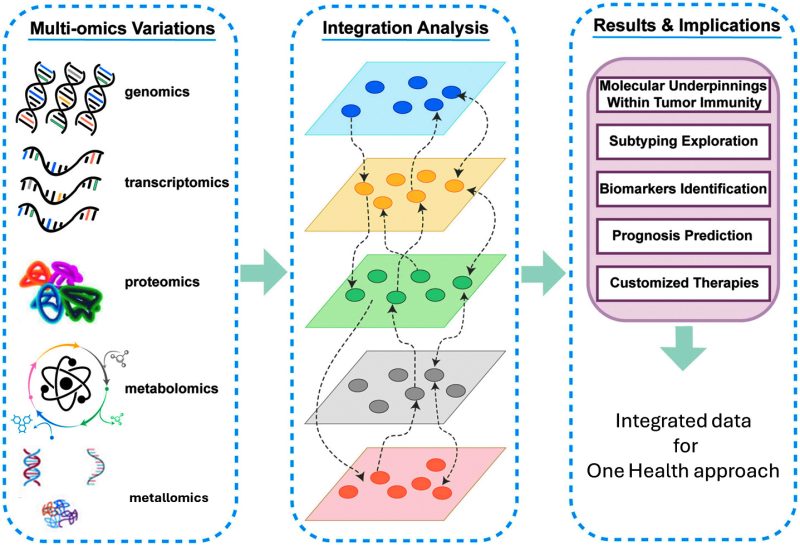
Fig. 5. Integrated multi-omics approaches. Available via license CC BY 4.0, adapted from [43].
In the context of One Health, Omics approaches are particularly relevant for controlling zoonoses, reducing bacterial resistance to antimicrobial agents, and ensuring food safety. They are useful for studying transmission pathways, pathogenic mechanisms, vaccine development, resistance mechanisms, and novel treatments for pathogens, toxins, and allergens. A challenging task in zoonosis control is the study of prion diseases to identify biomarkers for early diagnosis [44]. In this context, proteomics has been employed to clarify the molecular pathogenesis of prion diseases by studying protein-protein interactions, polymorphisms, and alterations in iron metabolism. The role of proteomics in One Health also includes studying antibiotic-resistant bacteria, which can cause zoonotic infections. Proteomics aims to elucidate the mechanisms through which bacteria develop resistance to antimicrobial agents and to evaluate novel treatments against such organisms. This expanding field is also integral to the food industry, facilitating the integration of processes related to animal health into food production. Proteomics technologies enable the monitoring of food production quality and the identification of potential threats to consumer health, such as the presence of enterotoxins in dairy products, the detection of allergens, and monitoring pathogens that could be transmitted to humans through food. Metabolomics involves the extensive study of small molecules that constitute the metabolome, reflecting the downstream effects of an organism’s genome and its interactions with the environment. As a relatively new discipline, metabolomics has broad applications in medical, clinical, biological, environmental, and agricultural sciences, aligning perfectly with the One Health approach [45]. While the application of metabolomics in veterinary medicine is not as widespread as in human medicine, its use is increasing, particularly due to its non-invasive nature and reliance on biological fluids. Metabolomics studies in these fields are crucial for preventing and controlling zoonotic diseases by understanding transmission mechanisms and developing nutritional interventions. This field will also guide the creation of nutraceuticals and functional foods tailored to an individual’s metabolic profile, enhancing therapeutic outcomes. Natural alternatives to antibiotics for both animal and human health can also be proposed. Additionally, metabolomics will assess the effects of environmental pollutants on metabolic health in humans and animals. In the domain of Omics approaches, metallomics is an emerging area. It integrates research related to metals in biological systems with genomics and proteomics by studying the interactions and functional connections of metal ions or species with genes, proteins, metabolites, and other biomolecules. Metallomics studies have predominantly focused on developing anticancer metal-based drugs and investigating cellular uptake assays of potential metallodrugs. Recently, Omics strategies have been proposed to unravel the mechanism of action of antiparasitic metal-based drugs. Currently, the frontier lies in integrating multidimensional metal analysis with spectroscopy and microscopic imaging for in situ metallomic imaging. The development of bioimaging workflows has the potential to advance our understanding of the roles of metals in biology [46]. Omics methods allow for comprehensive molecular target analysis, while microscopy techniques can be used to study the distribution of molecular targets, elucidating mechanisms such as disease development, transmission, and drug monitoring. Recent technological advancements supported by computational methods have made the direct correlation and integration of Omics approaches and imaging an exciting prospect.
5. Conclusions
To address the growing threat of emerging infectious diseases, zoonotic diseases, and antimicrobial resistance, it is essential to implement the One Health strategy and related interventions widely. Implementing a One Health approach can prevent diseases, reduce costs, enhance food safety and security, improve environmental monitoring, and save lives. For example, potential disease outbreaks can be detected early in animals, preventing their emergence and spread to human populations. Confronting these challenges and strengthening the mechanisms that facilitate global health management are crucial steps forward. This needs to be addressed by multidisciplinary collaborations and sustainable multidisciplinary solutions. Since the inception of the One Health approach, numerous interventions have been established across various regions and countries to address complex health issues, such as epidemics and pandemics. In the developed world, many collaborative platforms have been formed with an international strategy to address both environment-specific and health concerns. These strategies benefit significantly from analytical techniques that enable the rapid, low-cost, and early detection of markers related to human and animal health and the environment. Among these techniques, portable biosensors provide rapid and low-cost accurate analysis, while Omics approaches can identify an increasing number of targets and elucidate specific mechanisms. These emerging tools are instrumental in advancing the One Health agenda.
References
1. Zinsstag, J.; Schelling, E.; Crump, L.; Whittaker, M.; Tanner, M.; Stephen, C., eds. One Health: the theory and practice of integrated health approaches. 2nd edition, CABI Digital Library, 2020. https://www.cabidigitallibrary.org/doi/book/10.1079/9781789242577.0000.
2. Friedman, Y. Who is the biological patient? A new gradational and dynamic model for one health medicine. HPLS 2022, 44, 61. doi.org/10.1007/s40656-022-00540-9.
3. Schultz, M.; Rudolf Virchow. Emerg. Infect. Dis. 2008, 14, 1480-1481. doi.org/10.3201/eid1409.086672.
4. Sundberg, J.P.; Schofield, P.N. One medicine, one pathology, and the one health concept. J. Am. Vet. Med. Assoc. 2009, 234, 1530-1531. doi.org/10.2460/javma.234.12.1530.
5. Chien, Y.-J. How did international agencies perceive the avian influenza problem? The adoption and manufacture of the ‘One World, One Health’ framework. Sociology of Health & Illness 2013, 35, 213-226. doi.org/10.1111/j.1467-9566.2012.01534.x.
6. Zinsstag, J.; Schelling, E.; Waltner-Toews, D.; Tanner, M. From “one medicine” to “one health” and systemic approaches to health and well-being. Prev. Vet. Med. 2011, 101, 148-156. doi.org/10.1016/j.prevetmed.2010.07.003.
7. Martinengo, B.; Diamanti, E.; Uliassi, E.; Bolognesi, M.L. Harnessing the 12 Green Chemistry Principles for Sustainable Antiparasitic Drugs: Toward the One Health Approach. ACS Infect. Dis. 2024, 10, 6, 1856-1870. doi.org/10.1021/acsinfecdis.4c00172.
8. Johnson, M.S.; Adams, V.H. Integrating “One Health” Concepts in the Design of Sustainable Systems for Environmental Use. Toxics 2023, 11, 280-294. doi.org/10.3390/toxics11030280.
9. Gao, P. The Exposome in the Era of One Health. Environ. Sci. Technol. 2021, 55, 2790-2799. doi.org/10.1021/acs.est.0c07033.
10. Walker, D.I.; Valvi, D.; Rothman, N.; Lan, Q.; Miller, G. W.; Jones, D.P. The Metabolome: A Key Measure for Exposome Research in Epidemiology. Curr. Epidemiol. Rep. 2019, 6, 93-103.
11. Zhang, P.; Carlsten, C.; Chaleckis, R.; Hanhineva, K.; Huang, M.; Isobe, T.; Koistinen, V.M.; Meister, I.; Papazian, S.; Sdougkou, K.; Xie, H.; Martin, J.W.; Rappaport, S.M.; Tsugawa, H.; Walker, D.I.; Woodruff, T.J.; Wright, R.O.; Wheelock, C.E. Defining the Scope of Exposome Studies and Research Needs from a Multidisciplinary Perspective. Environ. Sci. Technol. Lett. 2021, 8, 839-852. doi.org/10.1021/acs.estlett.1c00648.
12. Jones, D.P. Sequencing the exposome: A call to action. Toxicol. Rep. 2016, 3, 29-45. doi.org/10.1016/j.toxrep.2015.11.009.
13. Escher, B.I.; Stapleton, H.M.; Schymanski, E.L. Tracking complex mixtures of chemicals in our changing environment. Science 2020, 367, 388-392. doi.org/10.1126/science.aay6636.
14. Maret, W. The quintessence of metallomics: a harbinger of a different life science based on the periodic table of the bioelements. Metallomics 2022, 14, mfac051. doi.org/10.1093/mtomcs/mfac051.
15. Liu, R.; Wu, P.; Yang, L.; Hou, X.; Lv, Y. Inductively coupled plasma mass spectrometry-based immunoassay: A review. Mass Spectrom. Rev. 2013, 5, 373-393. doi.org/10.1002/mas.21391.
16. Xuan, L.; Ju, Z.; Skonieczna, M.; Zhou, P.K.; Huang R. Nanoparticles-induced potential toxicity on human health: Applications, toxicity mechanisms, and evaluation models. MedComm 2023, 4, e327. doi.org/10.1002/mco2.327.
17. De Olivera Mallia, J.; Galea, R.; Nag, R.; Cummins, E.; Gatt, R.; Valdramidis, V. Nanoparticle Food Applications and Their Toxicity: Current Trends and Needs in Risk Assessment Strategies. J. Food Protection 2022, 85, 355-372. doi.org/10.4315/JFP-21-184.
18. Ferreira, I.; Garcia, O.; & Carnielli, L. Chapter 6 – The carbon footprint of cataract surgery and ISBCS. Immediately Sequential Bilateral Cataract Surgery (ISBCS): Global History and Methodology 2022, 75-83. doi.org/10.1016/B978-0-323-95309-2.00052-0.
19. ISO – ISO 14040:2006 – Environmental management – Life cycle assessment – Principles and framework. (n.d.). Retrieved December 29, 2020. https://www.iso.org/standard/37456.html.
20. Segerkvist, K.A.; Hansson, H.; Sonesson, U.; Gunnarsson, S. Research on Environmental, Economic, and Social Sustainability in Dairy Farming: A Systematic Mapping of Current Literature. Sustainability 2020, 12, 5502-5516. doi.org/10.3390/SU12145502.
21. FAO 2022. FAO Publications catalogue 2022 – October. Rome. https://doi.org/10.4060/CC2323EN.
22. Corcoran, J.; Winter, M.J.; Tyler, C.R. Pharmaceuticals in the Aquatic Environment: A Critical Review of the Evidence for Health Effects in Fish. Crit. Rev. Toxicol. 2010, 4, 287-304. doi:10.3109/10408440903373590.
23. Zanni, S.; Roccaro, M.; Bocedi, F.; Peli, A.; Bonoli, A. LCA to Estimate the Environmental Impact of Dairy Farms: A Case Study. Sustainability 2022, 14, 6028-6043. doi.org/10.3390/SU14106028.
24. Sambri, V.; Cavrini, F.; Rossini, G.; Pierro, A.; Landini, M.P. The 2007 epidemic outbreak of Chikungunya virus infection in the Romagna region of Italy: a new perspective for the possible diffusion of tropical diseases in temperate areas? New Microbiol. 2008, 31 (3), 303-304.
25. Bordi, L.; Carletti, F.; Castilletti, C.; Chiappini, R.; Sambri, V.; Cavrini, F.; Ippolito, G.; Di Caro, A.; Capobianchi, MR. Presence of the A226V mutation in autochthonous and imported Italian chikungunya virus strains. Clin. Infect. Dis. 2008, 47 (3), 428-429. doi: 10.1086/589925.
26. Caputo, B.; Russo, G.; Manica, M.; Vairo, F.; Poletti, P.; Guzzetta, G.; Merler, S.; Scagnolari, C.; Solimini, A. A comparative analysis of the 2007 and 2017 Italian chikungunya outbreaks and implication for public health response. PLoS Negl. Trop. Dis. 2020, 14 (6): e0008159. doi: 10.1371/journal.pntd.0008159.
27. De Carli, G.; Carletti, F.; Spaziante, M.; Gruber, C.E.M.; Rueca, M.; Spezia, P.G.; Vantaggio, V.; Barca, A.; De Liberato, C.; Romiti, F.; Scicluna, M.T.; Vaglio, S.; Feccia, M.; Di Rosa, E.; Gianzi, F.P.; Giambi, C.; Scognamiglio, P.; Nicastri, E.; Girardi, E.; Maggi, F.; Vairo, F. Outbreaks of autochthonous Dengue in Lazio region, Italy, August to September 2023: preliminary investigation. Lazio Dengue Outbreak Group; Lazio dengue Outbreak Group. Euro Surveill. 2023, 28 (44): 2300552. doi: 10.2807/1560-7917.
28. Zannoli, S.; Sambri, V. West Nile Virus and Usutu Virus Co-Circulation in Europe: Epidemiology and Implications. Microorganisms 2019, 7 (7): 184. doi. 10.3390/microorganisms7070184.
29. Fros, J.J.; Miesen, P.; Vogels, C.B.; Gaibani, P.; Sambri, V.; Martina, B.E.; Koenraadt, C.J.; van Rij, R.P.; Vlak, J.M.; Takken, W.; Pijlman, G.P. Comparative Usutu and West Nile virus transmission potential by local Culex pipiens mosquitoes in north-western Europe. One Health 2015, 31-36, doi.org/10.1016/j.onehlt.2015.08.002.
30. Sambri, V.; Capobianchi, M.; Charrel, R.; Fyodorova, M.; Gaibani, P.; Gould, E.; Niedrig, M.; Papa, A.; Pierro, A.; Rossini, G.; Varani, S.; Vocale, C.; Landini, M.P. West Nile virus in Europe: emergence, epidemiology, diagnosis, treatment, and prevention. Clin. Microbiol. Infect. 2013, 19 (8), 699-704. doi: 10.1111/1469-0691.12211.
31. Pierro, A.; Landini, M.P.; Gaibani, P.; Rossini, G.; Vocale, C.; Finarelli, A.C.; Cagarelli, R.; Sambri, V.; Varani, S. A model of laboratory surveillance for neuro-arbovirosis applied during 2012 in the Emilia-Romagna region, Italy. Clin. Microbiol. Infect. 2014, 20 (7), 672-677. doi: 10.1111/1469-0691.12436.
32. Sambri, V.; Capobianchi, M.R.; Cavrini, F.; Charrel, R.; Donoso-Mantke, O.; Escadafal, C.; Franco, L.; Gaibani, P.; Gould, E.A.; Niedrig, M.; Papa, A.; Pierro, A.; Rossini, G.; Sanchini, A.; Tenorio, A.; Varani, S.; Vázquez, A.; Vocale, C.; Zeller, H. Diagnosis of west nile virus human infections: overview and proposal of diagnostic protocols considering the results of external quality assessment studies. Viruses 2013, 5 (10), 2329-2348. doi: 10.3390/v5102329.
33. Halicka, K.; Cabaj, J. Electrospun Nanofibers for Sensing and Biosensing Applications–A Review. Int. J. Mol. Sci. 2021, 22, 6357-6381. doi.org/10.3390/ijms22126357.
34. Fortunati, S.; Giannetto, M.; Giliberti, C.; Bolchi, A.; Ferrari, D.; Locatelli, M.; Bianchi, B.; Boni, A.; De Munari, I.; Careri, M. Smart Immunosensors for Point-of-Care Serological Tests Aimed at Assessing Natural or Vaccine-Induced SARS-CoV-2 Immunity. Sensors 2022, 22, 5463-5478. doi.org/10.3390/ s22145463.
35. D’Agata, R.; Giuffrida, M.C.; Spoto, G. Peptide Nucleic Acid-Based Biosensors for Cancer Diagnosis. Molecules 2017, 22, 1951-1966. doi:10.3390/molecules22111951.
36. Fortunati, S.; Giannetto, M.; Giliberti, C.; Mattarozzi, M.; Bertucci, A.; Careri, M. Magnetic Beads as Versatile Tools for Electrochemical Biosensing Platforms in Point-of-Care Testing. Anal. Sens. 2024, 4, e202300062. doi.org/10.1002/anse.202300062.
37. Fortunati, S.; Giliberti, C.; Giannetto, M.; Bertucci, A.; Capodaglio, S.; Ricciardi, E.; Giacomini, P.; Bianchi, V.; Boni, A.; De Munari, I.; Corradini, R.; Careri, M. A highly sensitive electrochemical magneto-genosensing assay for the specific detection of a single nucleotide variation in the KRAS oncogene in human plasma. Biosens. Bioelectron.: X 2023, 15, 100404. doi.org/10.1016/j.biosx.2023.100404.
38. Roda, A.; Cavalera, S.; Di Nardo, F.; Calabria, D.; Rosati, S.; Simoni, P.; Colitti, B.; Baggiani, C.; Roda, M.; L. Anfossi. Dual lateral flow optical/chemiluminescence immunosensors for the rapid detection of salivary and serum IgA in patients with COVID-19 disease. Biosens. Bioelectron. 2021, 172, 112765. doi.org/10.1016/j.bios.2020.112765.
39. Domingo-Roca, R.; Lasserre, P.; Riordan, L.; Macdonald, A.R.; Dobrea, A.; Duncan, K.R.; Hannah, S.; Murphy, M.; Hoskisson, P.A.; Corrigan, D.K. Rapid assessment of antibiotic susceptibility using a fully 3D-printed impedance-based biosensor. Biosens. Bioelectron.: X 2023, 13, 100308. doi.org/10.1016/j.biosx.2023.100308.
40. Neethirajan, S. Recent advances in wearable sensors for animal health management. Sens. Bio-Sens. Res. 2017, 12, 15-29. doi.org/10.1016/j.sbsr.2016.11.004.
41. Erdem, A.; Eksin, E.; Senturk, Yildiz, E.; Maral, M. Recent developments in wearable biosensors for healthcare and biomedical applications. TrAC – Trends Anal. Chem. 2024, 171, 117510. doi.org/10.1016/j.trac.2023.117510.
42. Gavrilaș, S; Ursachi, C.Ș.; Perța-Crișan, S.; Munteanu, F.D. Recent Trends in Biosensors for Environmental Quality Monitoring. Sensors 2022, 22, 1513-1532. doi: 10.3390/s22041513.
43. Chen, S.; Xu, H.; Guo, C.; Liu, Z.; Han, X. The role of multi-omics variants in tumor immunity and immunotherapy. Front. Immunol. 2022, 13, 1098825. doi: 10.3389/fimmu.2022.1098825.
44. Silvia, C.J.; Erickson-Beltran, M. Detecting Differences in Prion Protein Conformation by Quantifying Methionine Oxidation. ACS Omega 2022, 7 (3), 2649-2660. doi.org/10.1021/acsomega.1c04989.
45. Hotea, I.; Sirbu, C.; Plotuna, A.-M.; Tîrziu, E.; Badea, C.; Berbecea, A.; Dragomirescu, M.; Radulov, I. Integrating (Nutri-)Metabolomics into the One Health Tendency–The Key for Personalized Medicine Advancement. Metabolites 2023, 13, 800. doi.org/10.3390/metabo13070800.
46. Stewart, T.J. Across the spectrum: integrating multidimensional metal analytics for in situ metallomic imaging. Metallomics 2019, 11, 29-49. doi.org/10.1039/C8MT00235E.
1 All images are reproduced in colour in the online edition of the Annales.


